

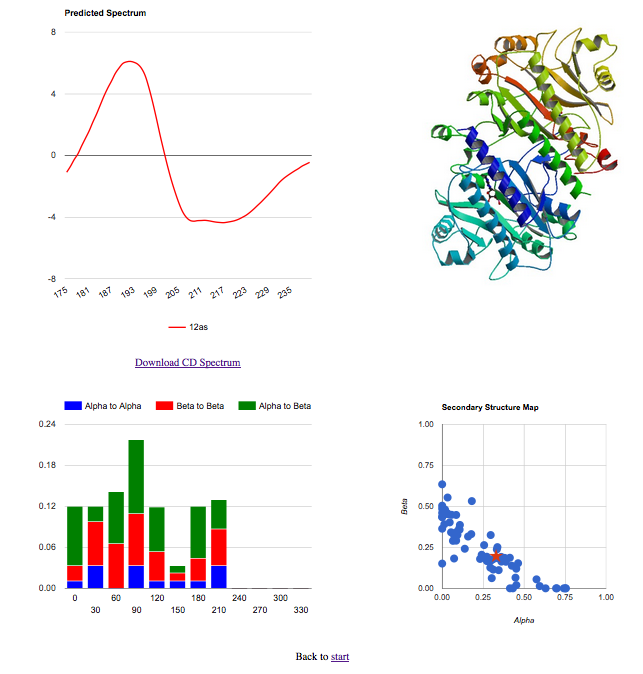
Server Input- Upload PDB: If you have your own pdb file, click on browse and upload your file to the server. - Give a PDB Code: Provide the 4 digits pdb code of the protein you want to predict. - (New) Reference Database: Choose from the SP175 (ref) or SMP180 (ref) SRCD reference databases. SP175 contains only soluble proteins and outputs predicted spectra in the range 175nm to 260nm. SMP180 contains all the proteins in SP175 with the addition of 30 membrane proteins, improving predictions for membrane proteins and many soluble proteins. SMP180 outputs predicted spectra in the range 180nm to 260nm. Result PageThe result page is split into four panes:- Top Left: The visual representation of the predicted spectrum, and a link to a file with a two column format. - Top Right: (Optional) If the user has checked against a pdb code, the server will download the image of the protein from RCSB, which is shown. In case the user uploads his own file, instead of the protein visualization the bottom right pane is shown in this position. - Bottom Left: The topology of the pdb structure is shown, all the secondary element connections grouped according to three types alpha to alpha, alpha to beta and beta to beta. - Bottom Right: The secondary structure map of the SP175 data set is shown as blue dots on the scatter plot and the input protein as a red star. The reference database used is indicated at the top of the results page and in the downloadble CD spectra text file. The graphs can be downloaded as .png files for your records by right clicking the "Downloadable version" link and selecting "Save link as..." in the pop up menu. |
|
CiteIf you use PDB2CD, please quoteIn using PDB2CD please cite: Mavridis, L,. and Janes, R.W.. (2017) PDB2CD : A web-based application for the generation of circular dichroism spectra from protein atomic coordinates. Bioinformatics 33: 56-63.
|
ContactDr. Robert W. Janes - r.w.janes at qmul.ac.uk Elliot D. Drew - e.drew at qmul.ac.uk |

|

|

|